NBHF
Methods Summary
Pamguard v.2.02.09 (https://www.pamguard.org/) is used to run click detector on full bandwidth data (384 kHz sample rate)
Data is high-pass filtered at 100 kHz with a click detector (12 dB threshold)
Pamguard v2.02.09f (public release expected to be available in early 2024) is used to reclassify clicks
Click detections are classified within Pamguard based on peak frequency
The Matched Template Classifier module evaluates the similarity of each click detection to known templates
Potential acoustic events are automatically defined in R
PAMpal is used to calculate features of click detections in each event
Step-by-Step Instructions
Run echolocation click detector
Define location to save database and binary files
Define array configuration (hydrophone sensitivity and separation)
Save configuration file with a descriptive filename
Press Run
Reclassify clicks with click classifiers and matched template classifier
- Import Matched Template Classification module and Click Detector from NBHF_ClickClassifiers_and_MTC.psfx found in content/ToothedWhales/Pamguard settings directory
Click classifiers are saved within the Click Detector module in the default NBHF_ClickClassifiers_and_MTC configuration file. They are also saved as individual .pgccs files within content/ToothedWhales/Click Classifiers/NBHF directory.
In the Pamguard File Menu, select “Import PAMGuard Modules”
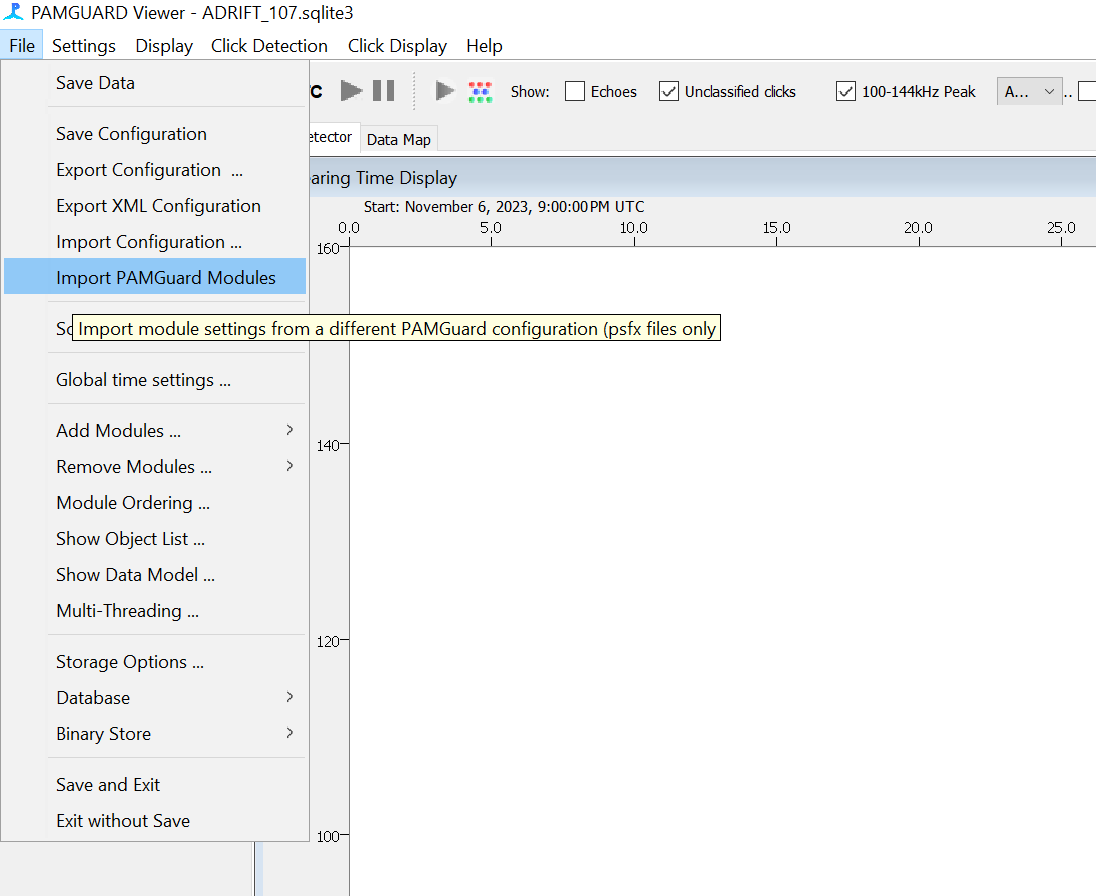
For Click Detector, select “Replace existing Click Detector” and for Match Template Classifier, select “Import as new module”
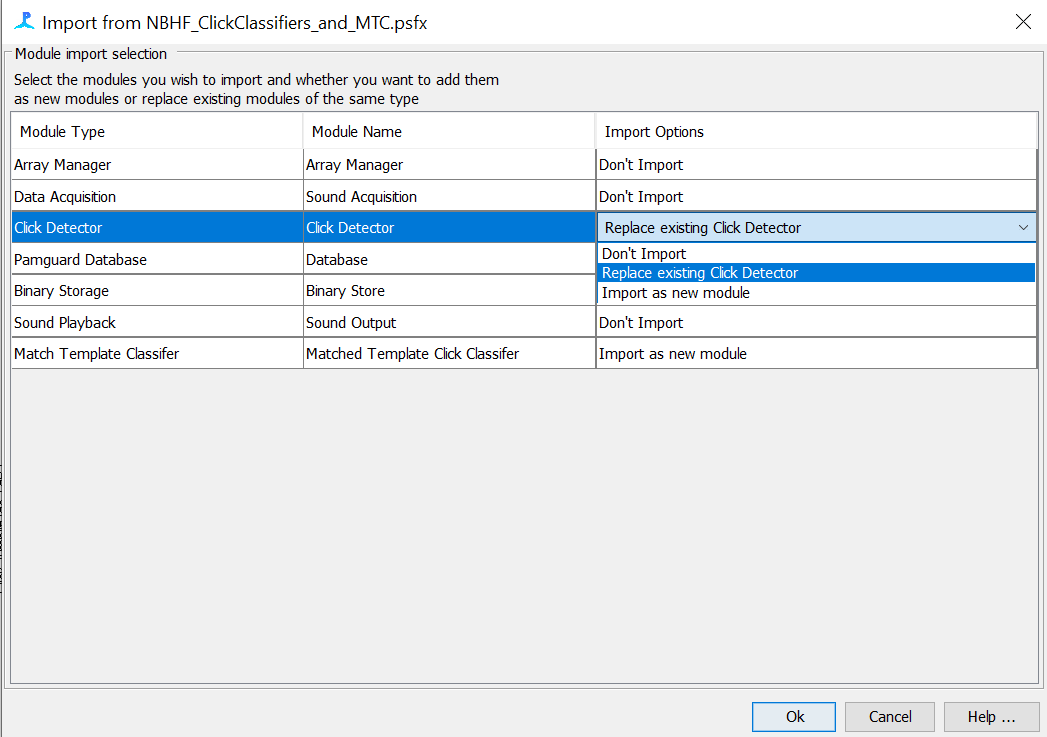
- Reclassify clicks with NBHF click classifiers
In the Pamguard Click Classifier menu, select “Reanalyse Clicks”
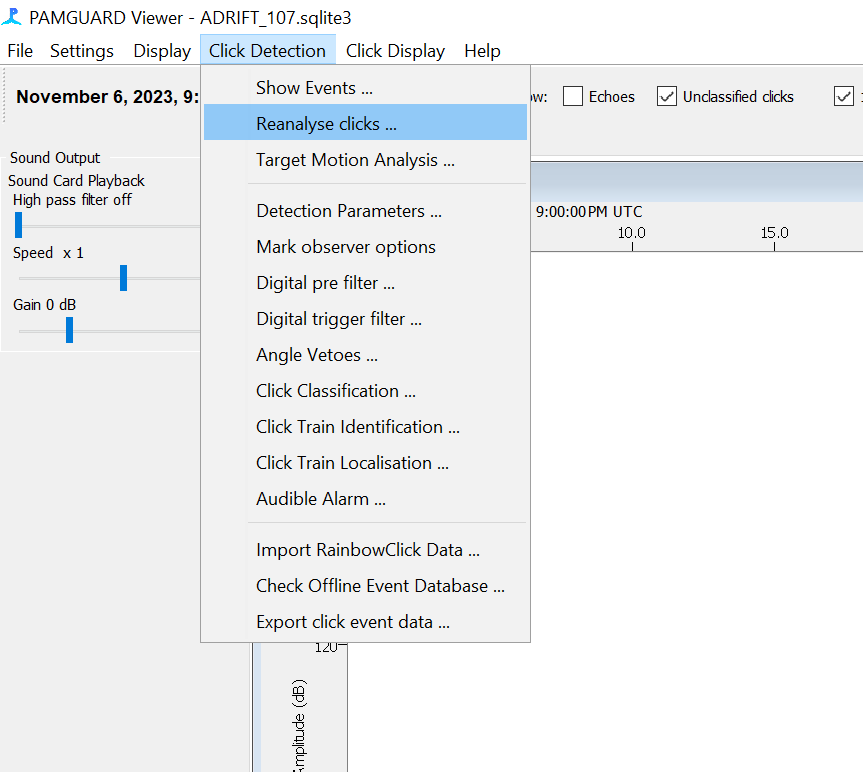
Select “All Data”, tick the “Delete old database entries” box, and tick the “Reclassify Clicks” box. Add a note “Reclassify based on NBHF peak frequencies”. Then click “Start”
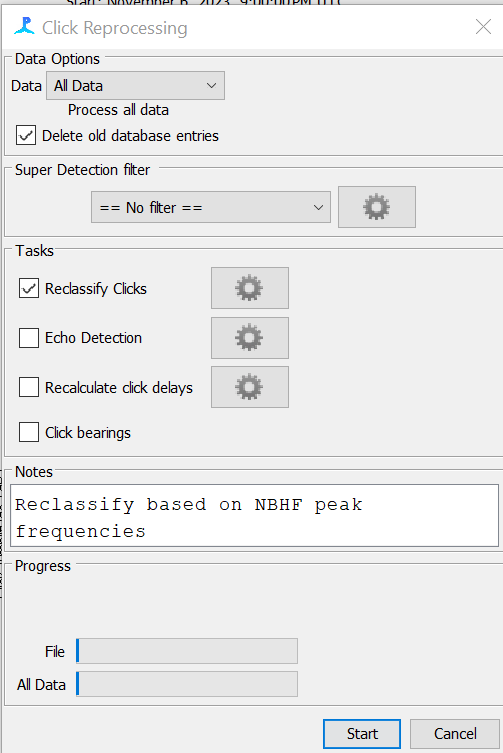
- Reclassify clicks for Matched Template Classifier
In the Pamguard Settings menu, select “Matched Template Classifier” and “Reclassify clicks”
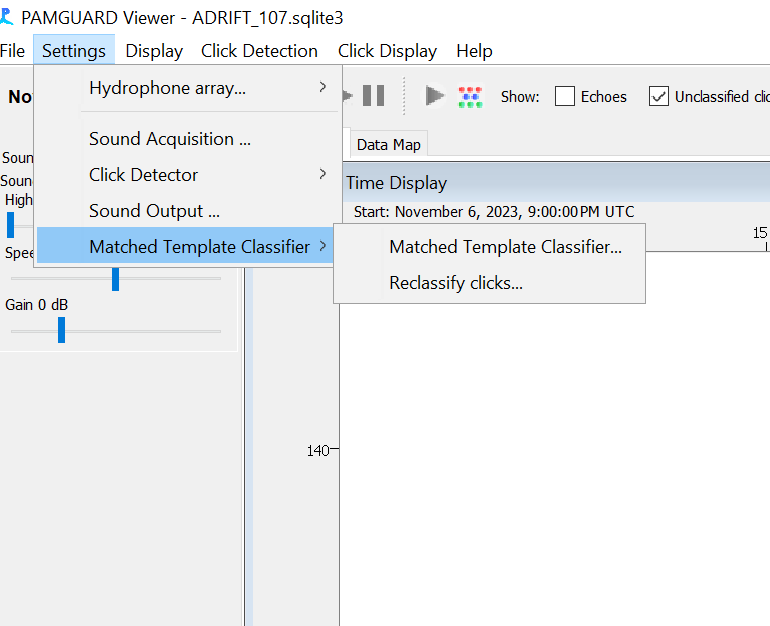
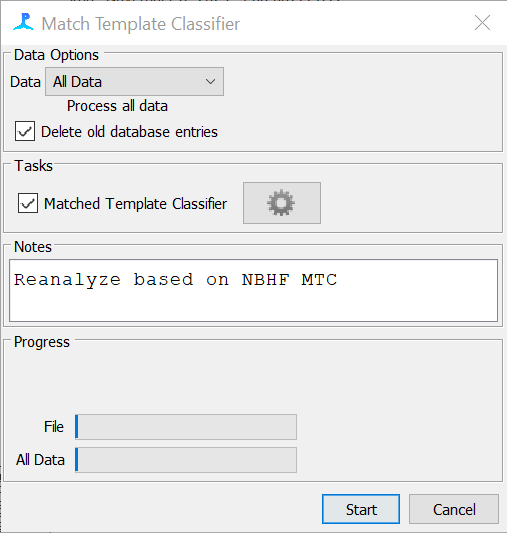
Select “All Data”, tick the “Delete old database entries” box, and tick the “Matched Template Classifier” box. Add a note “Reanalyze based on NBHF MTC”. Then click “Start”
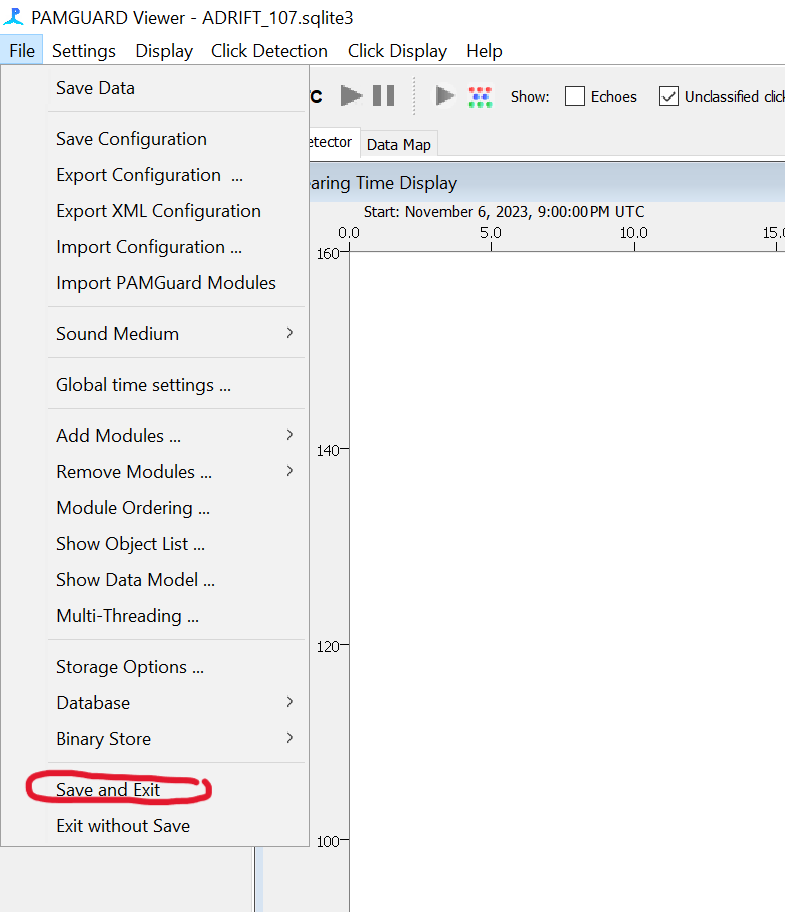
- When finished, save data and close Pamguard
Automatically define potential acoustic events in R
Four thresholds are used to identify potential NBHF events, including “Pd_1” and “Pd_2” for Dall’s porpoise, “Pp” for Harbor porpoise, and “Ksp” for Kogia. Template thresholds: Pd_1=0.45, Pd_2=0.45, Pp=0.6, Ksp=0.45. These click templates were generated from acoustic encounters with known species. Thresholds were established in an iterative process to ensure no high quality acoustic events were missed.
The matchTemplateScript_NBHF.R script is used to identify potential NBHF events, add them to a database, and then generate an acoustic study using the PAMpal R package.
Events are automatically defined when the following conditions are met
- 3 or more clicks exceed the MTC thresholds within 120 seconds
- Events have a maximum duration of 120 seconds